Organization and structure
#Organization Principle
The core principle of the ARC is that collected data are stored in directories, while metadata are maintained in accompanying tables that reference and describe the data. This organizational structure is closely aligned with the ISA model, while also incorporating workflows, computational processing, and analysis results.
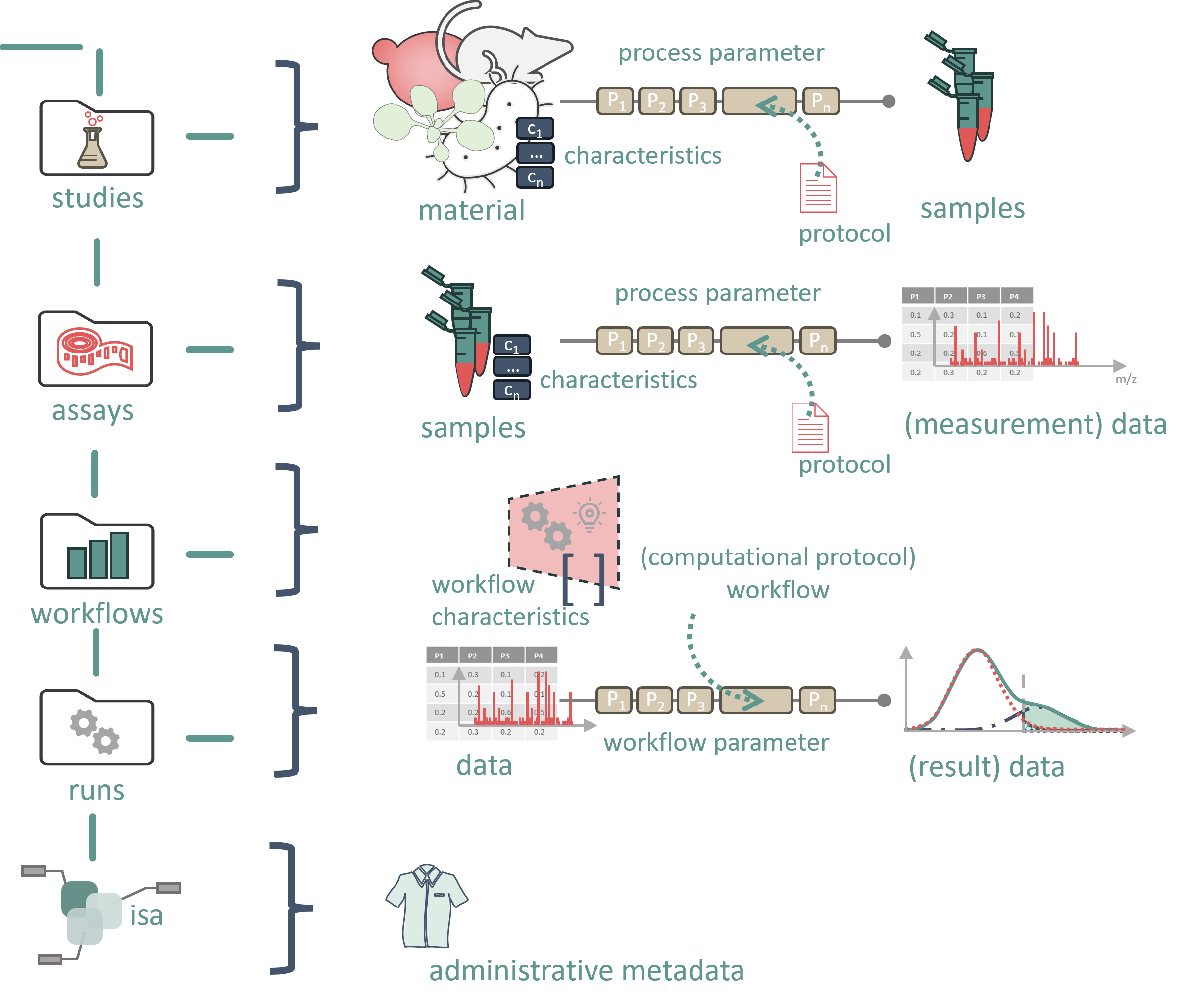
The foundational idea behind ARC is to provide a directory scaffold that ensures research data, along with their processing and analysis, are organized in a structured and annotated manner. This scaffold supplies a basic file structure for organizing research locally on a personal machine, as well as on data-producing devices such as measurement instruments or compute servers. A key feature of this system is its ability to seamlessly transfer to the cloud, specifically to a DataHUB instance (e.g., Git -LFS) hosted by an institution or NFDI consortium.
Through GIT ’s versioning mechanism, data can be easily backed up, integrated across devices, and shared with collaborators. Each interaction is tracked and can be reverted if necessary. Additionally, DataHUB offers project management tools, such as task assignments and discussion boards.
The unified structure of ARC ensures that research can be shared and understood easily by others. Several software tools are available to help create ARCs and support data analysis functionalities. While ARC requires a specific directory structure to be recognized, researchers have the flexibility to add additional files or folders as needed.
#ARC Directory Structure
ARC represents an entire investigation. At the top level, it includes directories named “studies,” “assays,” “workflows,” and “runs,” along with an investigation metadata table that holds all administrative metadata.
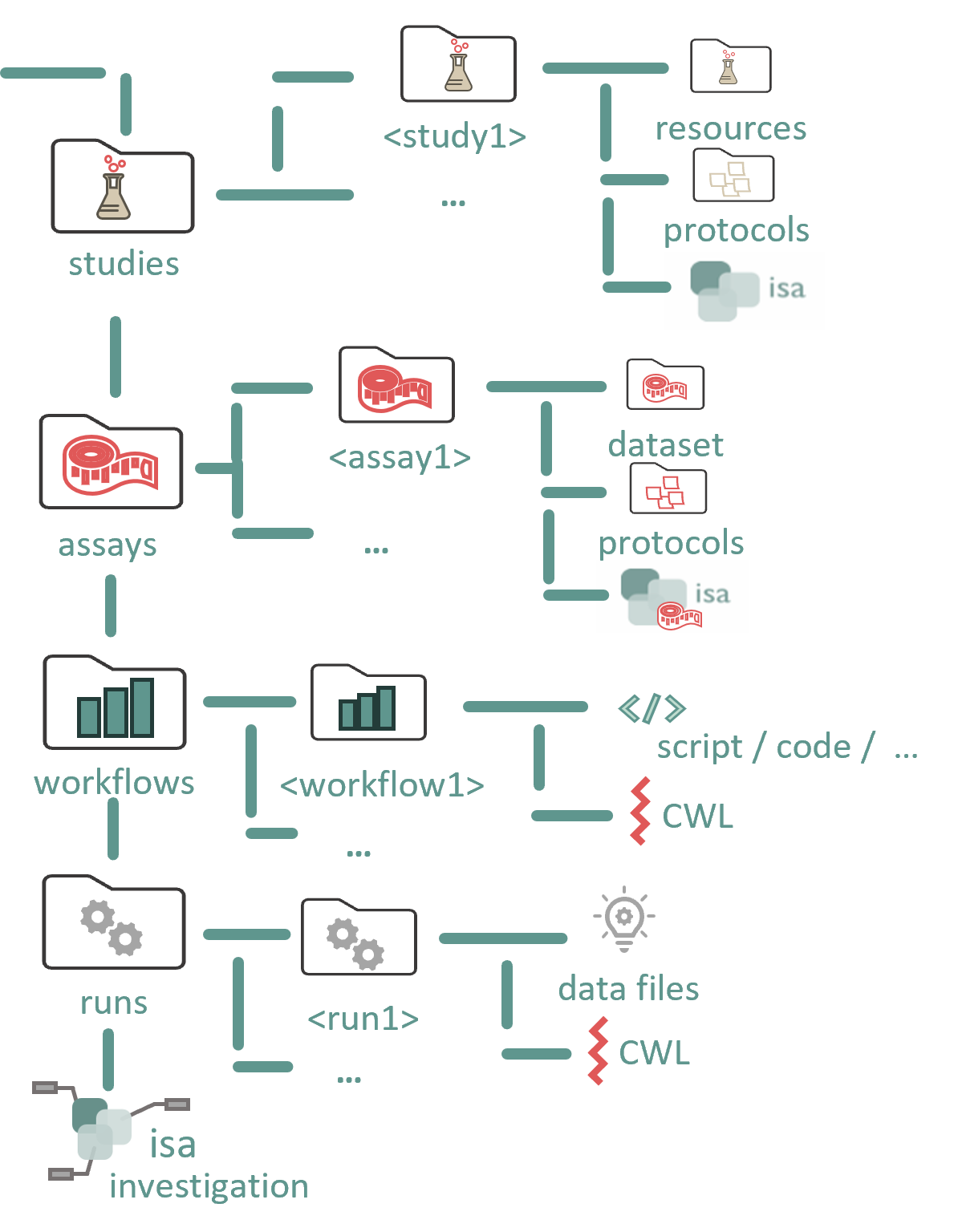
#Study
The studies folder contains one or more studies, each in its own directory. Each study folder contains:
- Study metadata file: A table with metadata describing the study.
- Resources folder: Contains external data used in the study.
- Protocols folder: Stores protocols that describe the process from starting material (or data) to samples. These protocols should be stored in a format that can be referenced in the metadata table.
#Assay
The assays folder contains one or more assays, each in its own directory. Each assay folder contains:
- Assay metadata file: A table with metadata describing the measurement process.
- Dataset folder: Holds the resulting data from the assay process, typically raw measurement files (open file formats are encouraged).
- Protocols folder: Contains the protocols that describe the process from samples to measurement.
#Workflows
The workflows folder contains subfolders for each workflow, which may include anything from simple scripts to full programs or toolchains for simulations, processing, or analysis. These workflows should not be tied to specific input/output files, which are instead managed by the metadata in a run.
- Workflow metadata file: Describes the executables and computational environment needed for the workflow.
#Runs
For each computational run, a separate folder is created to store the resulting data.
- Run metadata file: Contains specific parameter values for the run, including the input data used.
#Flexibility and Expansion
The ARC scaffold provides a well-defined space for organizing research data but does not require every aspect to be filled. Researchers and collaborators can use this structure as needed, leaving any irrelevant sections empty. Additional folders can be created for other research elements, such as paper drafts or notes, allowing for flexible expansion.
For more detailed information, refer to the [ARC Scaffold Specification].
#Continuous Quality Control
ARC supports continuous quality control in the background, ensuring data integrity without interrupting work. The entire investigation can be compiled into a data publication, assigned a DOI , and referenced in journal publications.